DEVELOPING eDNA AS TOOL FOR MONITORING CULTURED AND WILD POPULATIONS OF SEA SCALLOPS Placopecten magellanicus IN MAINE, USA
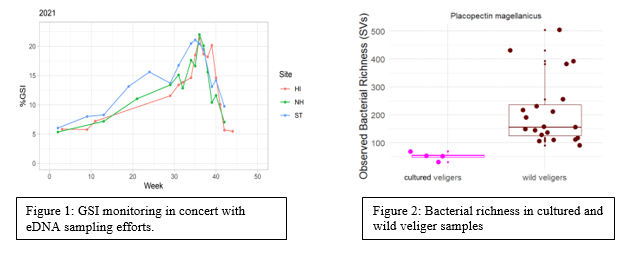
The Maine eDNA program aims to use molecular tools to better understand and promote sustainable use of wild and cultured resources. Here I will talk about two applications of eDNA tools for evaluating sea scallop aquaculture efforts along the coast of Maine, in the field, farm, and lab. One goal is to groundtruth eDNA assays for use in aquaculture in hatchery and field conditions. Quantitative eDNA assays for sea scallops have been developed and calibrated for sperm and dockside conditions, but this assay has not been calibrated for eggs or larvae, nor has it been tested in the field over wild beds and on functioning aquaculture farms. Here we explore whether eDNA from adults vs gametes and larvae can be distinguished by sampling at different depths and points in time in cultured and wild populations of sea scallops along the coast of Maine. In concert with traditional techniques like gonadal somatic indices (GSIs) (Figure 1), water samples were collected at three scallop aquaculture farms and three sites distant from farms in Penobscot Bay, ME, to assess the relationship between eDNA and spawning over time and magnitude.
In our next application we’re exploring how microbiomes (collections of microbes associated with scallops) vary over life stage in wild and cultured scallops and how this might influence hatchery success. Microbiomes of wild veligers from Southern Maine and cultured veligers from the Downeast Institute were surveyed using 16S metabarcoding and compared to microbiomes from tank water and biofilms. Bacterial communities from these different sources were compared to identify potentially pathogenic versus healthy bacteria to better understand the high failure rate of hatchery-reared scallop larvae (Figure 2).
Both of these studies highlight how eDNA can be applied to improve aquaculture development, operations and management. We’ll discuss the results of our studies and also the general process of eDNA assay development and processing for commercial applications.