USING eDNA METABARCODING TO CHARACTERIZE MARINE FISH ASSEMBLAGES AND MICROBIAL COMMUNITIES
eDNA metabarcoding, designed to characterize complex communities, is being applied to answer a wide range of resource management questions by the NOAA Northeast Fisheries Science Center. Finfish communities were described over 3 years using eDNA from sampled water near oyster farms in Long Island Sound (Fig 1,doi:10.3389/fmars.2019.00674 ). Microorganisms growing on aquacultured
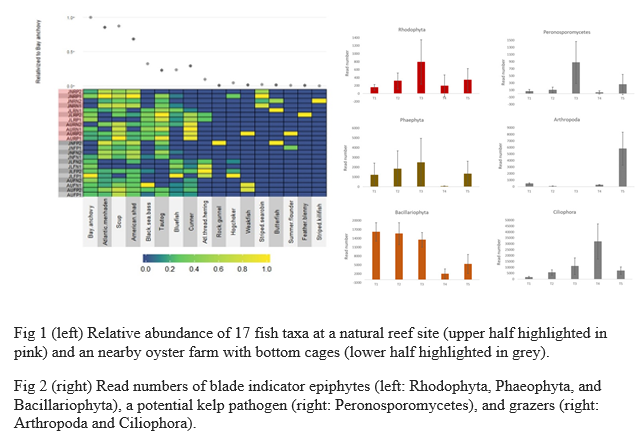
sugar kelp were compared with those in surrounding seawater, and indicator species important to sugar kelp ecology and health were identified (Fig 2,doi:10.1016/j.algal.2022.102654 ). A pilot project analyzed eDNA in seal feces to identify fish consumed by grey seal and harbor seal. A NOAA Fisheries Strategic Initiative is using ongoing eDNA sampling over the northeastern continental shelf to advance this technology for living marine resource assessment. T hese projects are summarized and synthesized with respect to the potential benefits and challenges of using eDNA for the management of living marine resources.