USING MICROBIAL GENOMICS TO MONITOR ECOSYSTEM RESPONSES TO ANTHROPOGENIC PERTURBATIONS: OIL SPILLS AS A CASE STUDY
Microbial communities form the base of the food webs in the ocean, with phytoplankton fixing CO2 to produce biomass, microzooplankton consuming this biomass and repackaging it for larger consumers and Bacteria and Archaea, “Prokaryotes”, recycling and repackaging waste products. These small organisms have short generation times, are abundant and contribute to a wide range of biogeochemical cycles. These characteristics make microbes ideal organisms for monitoring ecosystem responses to anthropogenic perturbations in the oceans.
Over the last several decades, advances in genomics technologies have led to the wide-spread use of amplicon sequencing in characterizing and monitoring microbial community composition and structure. Using primers designed to target a broad range (‘universal) of organisms, or to specifically target a small taxonomic group or functional genes, amplicon sequencing can provide detailed information about which organisms are present or which metabolic pathways or biogeochemical transformations are possible. While DNA is the most common biomolecule targeted in amplicon surveys, RNA can also be targeted, providing information about active microbes and pathways.
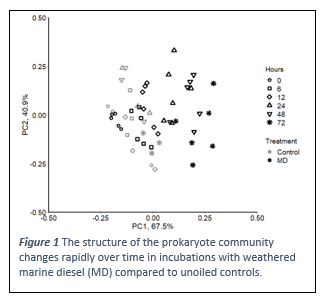
Microbes have long been known to be active players in the recovery of marine ecosystems following an oil spill. Several genera of Bacteria, some fungi and a few phytoplankton have been identified as being able to degrade hydrocarbons in oil. Other groups may respond positively or negatively to oil, with responses occurring within hours or a few days. This rapid response may be useful in monitoring for hydrocarbon pollution but can also provide information about the fate and likely impacts of oil following a spill. Using a series of mesocosm experiments, ranging from 12 to 215 L, we have quantified the response of microbial communities to diluted bitumen, diesel and a medium crude using 16S rRNA amplicon sequencing. The microbial communities respond rapidly to hydrocarbons (Figure 1), with similarities across oil types. Seasonal and geographical differences in the responses are generally a function of the initial microbial community, which reflects the local environmental conditions. Generally, microbes are capable of contributing to oil spill response, aiding recovery of ecosystems after a spill.