IDENTIFYING ISOFORM SWITCHES IN ATLANTIC SALMON Salmo salar INFECTED WITH Moritella viscosa
Alternative splicing is a regulatory process during gene expression that results in a single gene coding for multiple proteins. A vast majority of eukaryotic genes use alternative transcript isoforms. The alternative isoforms are generated through alternative transcription start sites (aTSS), alternative transcription termination sites (aTTS), and alternative splicing (AS). Gene isoforms are predominantly tissue-specific and may alter the stability, cellular localization and function of the gene. A switch is defined as a case where the relative contribution of the isoforms to the parent gene expression changes significantly between conditions (e.g. healthy vs. diseased). In this study, the skin transcriptome using RNA-seq of Atlantic salmon infected with Moritella viscosa (a Gram-negative bacterium that causes winter ulcer disease) was used to examine the isoform usage of immune genes in healthy vs. infection samples. The skin samples were collected from control and infected fish [at lesion (at) and away from lesion (adj) sites].
HISAT and StringTie software performed alignment and quantification of the skin transcriptome. IsoformSwitchAnalyzeR Bioconductor R package was used to analyze isoform usage, providing a platform to integrate many sources of predicted annotation such as open reading frame, protein domain (via Pfam), signal peptides (via SignalP), intrinsically disordered regions (IDR, via NetSurfP-2), coding potential (via CPAT or CPC2) and sensitivity to non-sense mediated decay (NMD). Differential isoform usage analysis was performed using DEXSeq and DRIMseq.
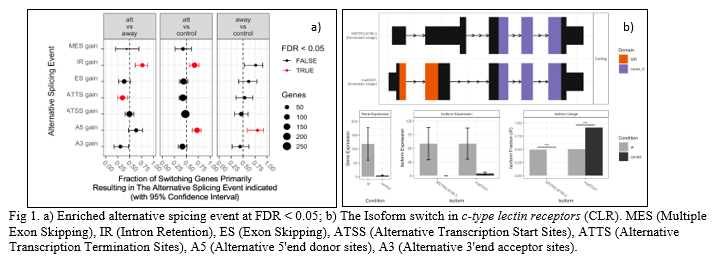
In total, 615 switches from 853 isoforms (486 genes) were identified. The comparison at lesion (at) and adjacent to lesion (adj) of infected fish with control (non-infected skin samples) resulted in 470 and 103 switches from 675 isoforms (383 genes) and 136 isoforms (986 genes), respectively. The genome-wide analysis of alternative splicing was enriched in intron-retention (IR gain), alternative transcription termination sites (ATTS gain) and alternative 5’end donor sites (A5 gain) at false discovery rate (FDR) < 0.05 (Fig. 1a). Some of the examples of enriched alternative isoform usage events were observed in cadherin-13, g0/g1 switch protein 2, and c type lectin receptor b. Signalling triggered by C-type lectin receptors (CLRs) plays crucial roles in activation of immune response against infections. CLRs have diverse immunoregulatory functions associated with phagocytic, anti-microbial, and inflammatory responses (Fig. 1b). Isomer switches identified in the present study may be employed as functional biomarkers to predict functions that impact vital biological processes.