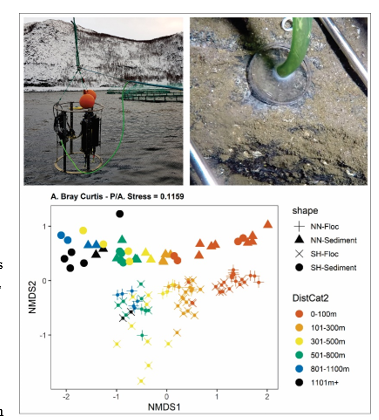
INTRODUCING SIBS: ‘SUBSTRATE INDEPENDENT BENTHIC SAMPLER’ FOR HARD AND MIXED-BOTTOM HABITATS: A PROOF OF CONCEPT STUDY
As is the case in most countries, the Norwegian standard for monitoring of organic impacts of salmon farming is centered
around the analysis of chemical and biological parameters of soft-sediments habitats . However, m any fish farms are situated in areas with hard or mixed substrates and this is likely to increase given the anticipated
shift to more physically dynamic sites.
Although a nalogous changes are probably occurring on these mixed-bottoms
habitats , our knowledge of
any
potential interactions is severely lacking , and more critically,
are going largely unmonitored due to the absence of a suitable sampling method.
There are however emerging alternative biological indicators that may provide a solution. Recently, several studies have demonstrated
the potential of using genetic material from micro-organisms (e.g. bacteria, foraminifera etc.), assessed
through rapid, high-throughput sequencing techniques, to discern benthic enrichment. To date, these have been necessarily restricted
to sampleable soft-sediment habitats. The small, ubiquitous, highly abundant and diverse nature of these
organisms means that they are likely to be present in the biofilms and/or flocculent layers that
overly most hard- and mixed-bottom substrates.
Here we present the first set of results using eDNA to describe the microbial communities of samples collected using a newly developed ‘Substrate Independent Benthic Sampler’ device. Samples were obtained from sand, shellsand , gravel, and bedrock using a low-cost surface operated suction device and are compared to more conventional microbial eDNA samples from soft-sediments and to macrofauna samples, where feasible. Minor differences were observed between sample types and farms, but in general, very g ood congruence wa s observed between the datasets with respect to distance from farm and anticipated benthic effects. Sampling of flocculent material overlying all substrates provides a potentially vital monitoring tool for the management of fish farm effects in the future.