GENETIC CHARACTERIZATION OF HETEROLOGOUS Edwardsiella piscicida ISOLATES FROM DIVERSE FISH HOSTS AND VIRULENCE ASSESSMENT IN A CHINOOK SALMON Oncorhynchus tshawytscha MODEL
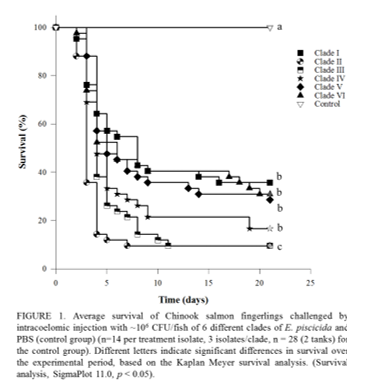
Edwardsiella piscicida is an emergent pathogen of finfish aquaculture. Although it is recognized as a pathogen with a wide host range, host associations driving genetic diversity remain unclear. This study investigated the genetic and virulence diversity of 37 E. piscicida isolates recovered from 10 fish species in North America. Multilocus sequence analysis (MLSA) was conducted using concatenated alignments of the gyrB, pgi and phoU sequences. MLSA clustered the tested isolates into six discrete genetic groups. In light of recent disease outbreaks in cultured salmonids, the virulence of each clade was evaluated in Chinook salmon Oncorhynchus tshawytscha fingerlings following intracoelomic challenge of ~106 CFU/fish. Challenged and control fish were monitored for 21d and microbiological and histological examination was performed on dead and survivor fish. Peak mortality occurred 3-5 days post-challenge (dpc) regardless of isolate or genetic group. Edwardsiella piscicida was recovered from all moribund and dead animals. At 21dpc, fish challenged with isolates from clades II, III and IV presented cumulative mortality ≥83. 3%, whereas isolates from clade I, V and VI resulted in cumulative mortality ≤71.4% (Figure 1). This study suggests an underlying genetic basis for strain virulence and potential host associations. Further investigations using other fish models and variable challenge conditions are warranted.